※ User Guide:
Frequently Asked Questions:
1 . Q: How to read the PULPS results?
A: Here we use the human protein FUS as the example. After clicking "Submit", the prediction results of its propensity to drive phase separation are shown as follows:
<1>. The table of the PULPS basic prediction results
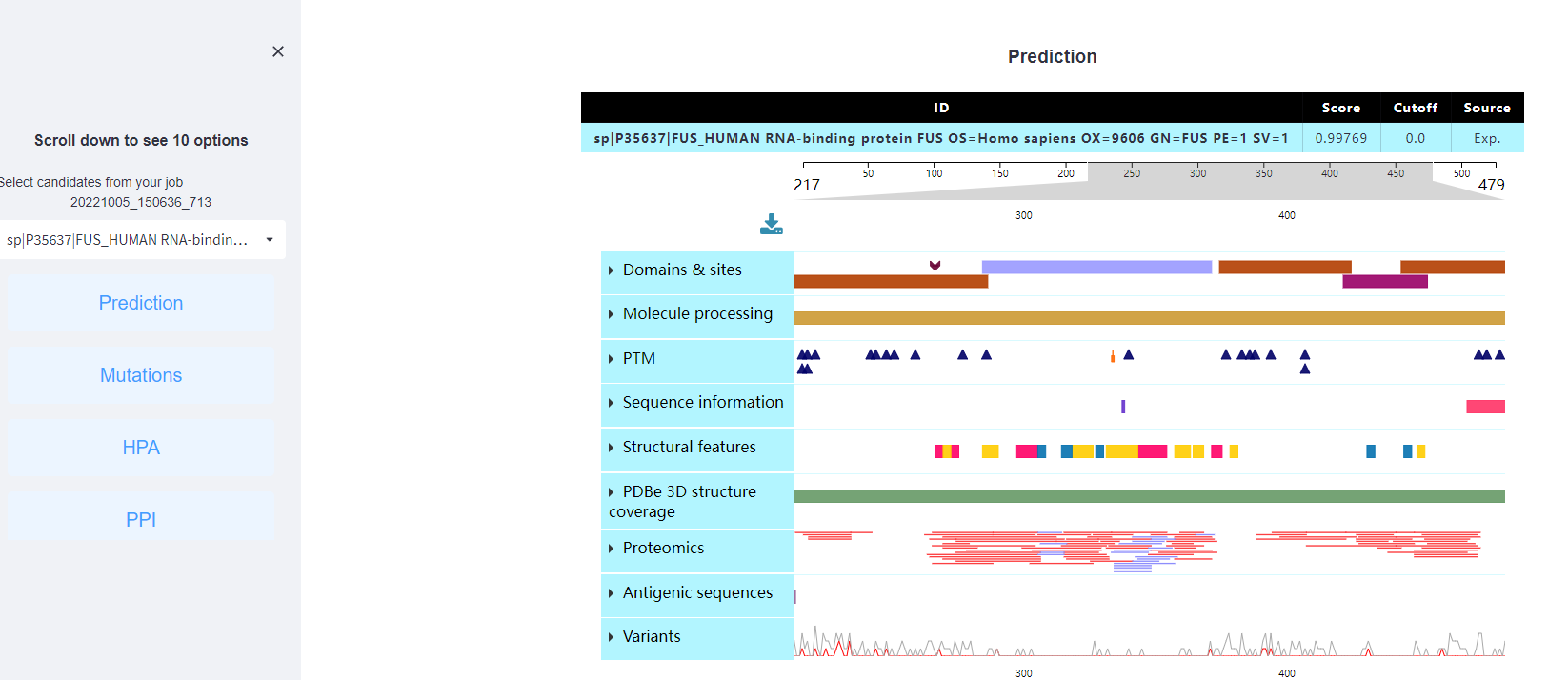
Score: The value calculated by PULPS to evaluate the potential of scaffolds in LLPS.
Cutoff: The cutoff value under the threshold. Different threshold means different precision, sensitivity and specificity. In this project, there is no specific cutoff.
Source: Whether the interactions is validated by experiment, "Exp." means YES, while "Pred." means NO.
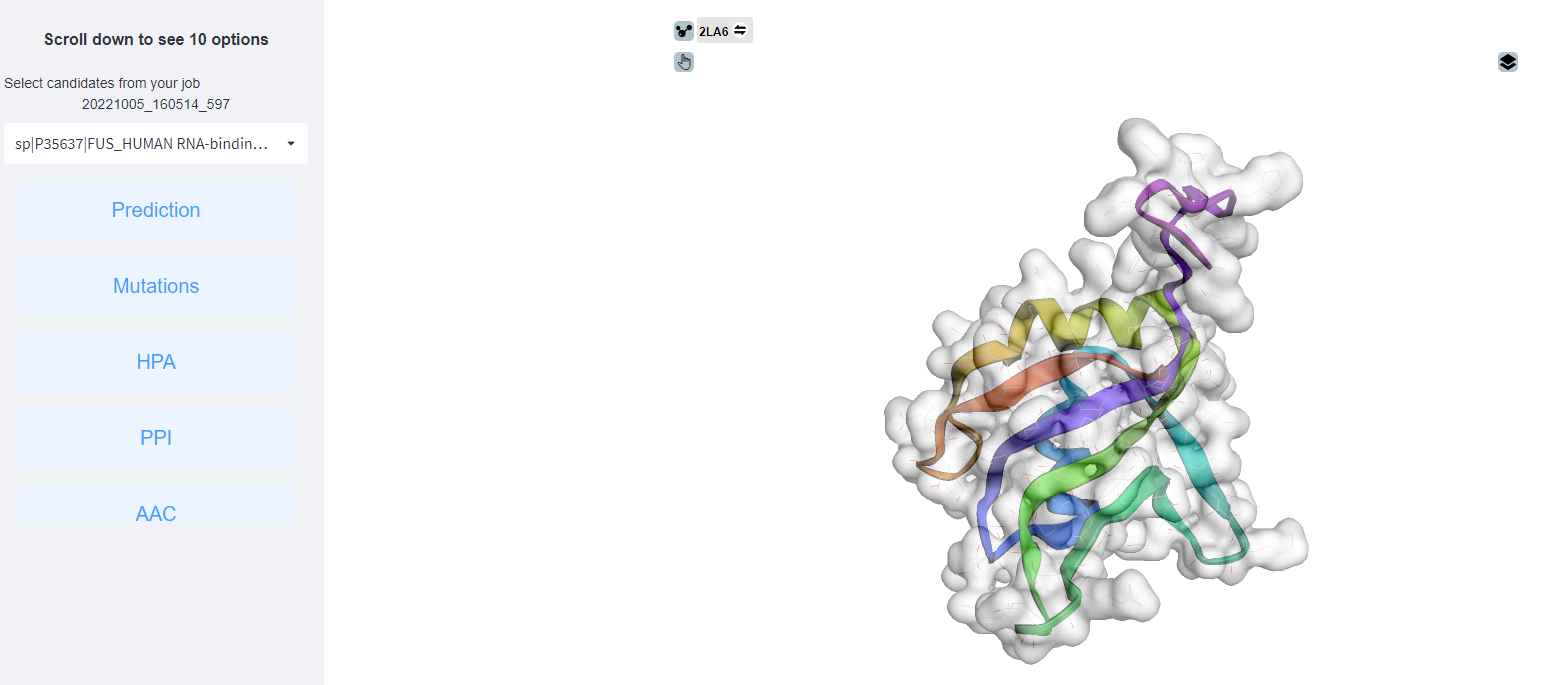
<2>. The results of the PDB structure.
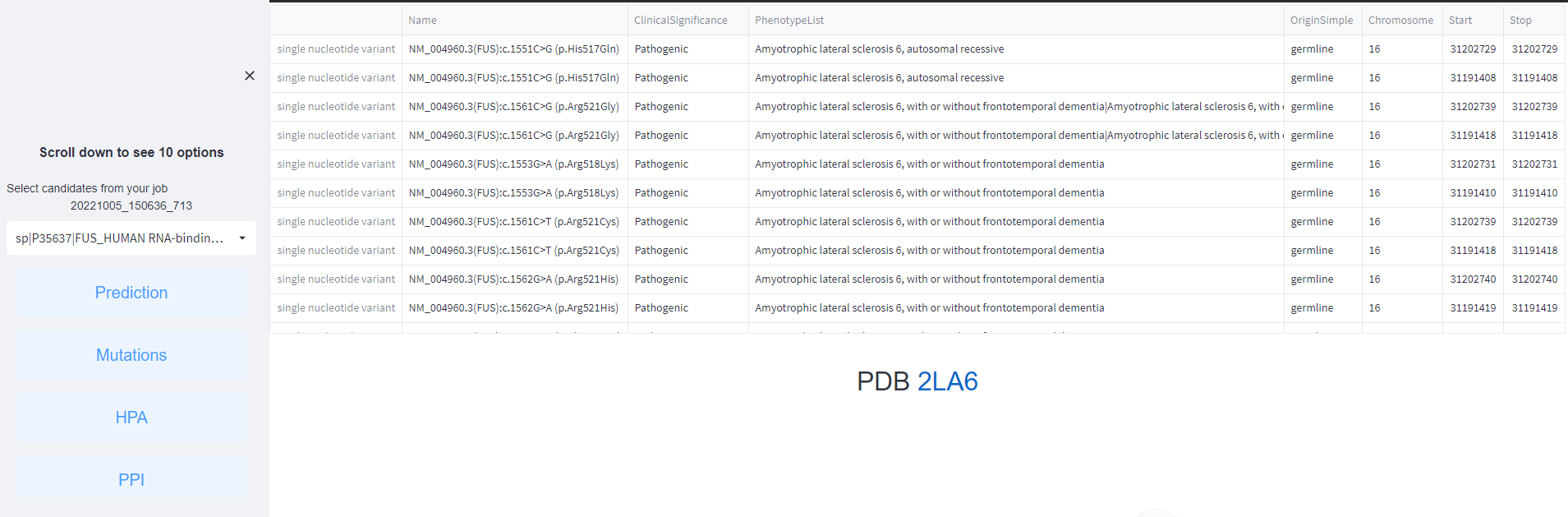
2. Q: How many protein features you use to characterize both scaffold and unlabeled proteins in LLPS in the human proteome?
A: Six types of protein features, including sequence-derived features along with amino acid composition, biophysical principles of the propensity towards droplet formation, IDR, hydrophobic regions, LCR, and secondary structure. Users can scroll down to see 10 options in the server page and browse by each feature results as mentioned above, users will get the informations of feature results presented in table pr picture format. For example, all features of protein FUS like the figure shown in below:
<1>. The results of Disease associated-mutations.
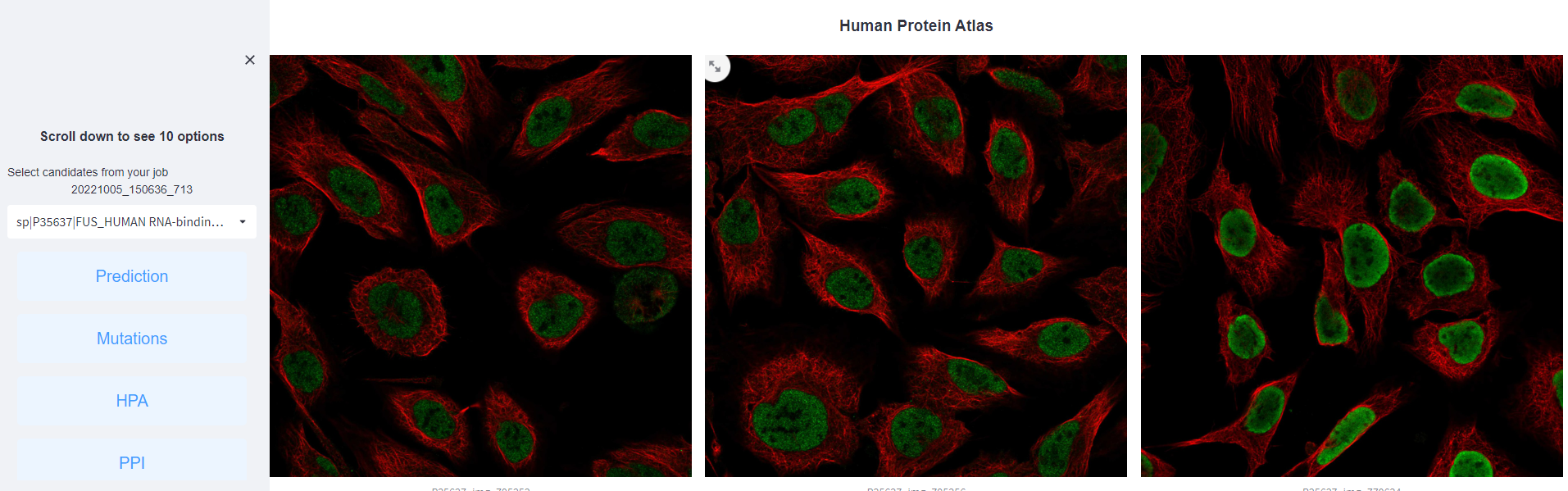
<2>. The results of Human Protein Atlas.
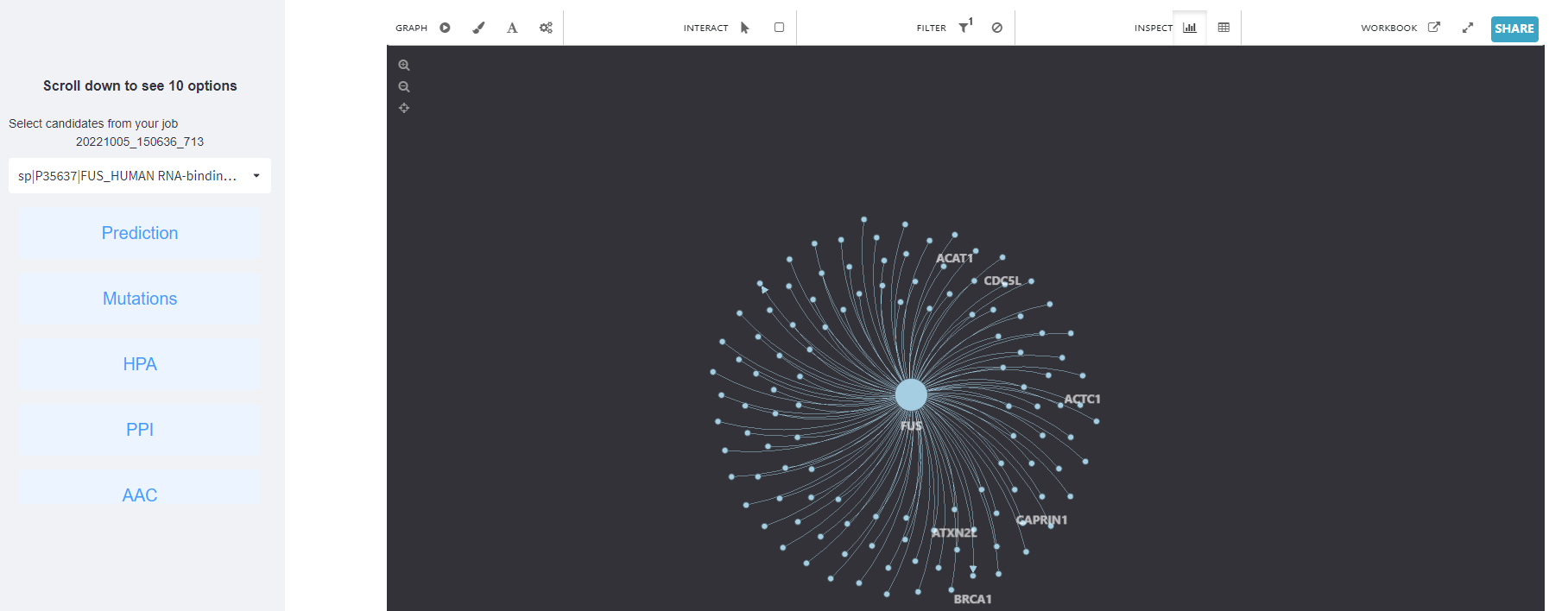
<3>. The results of Protein protein interaction.
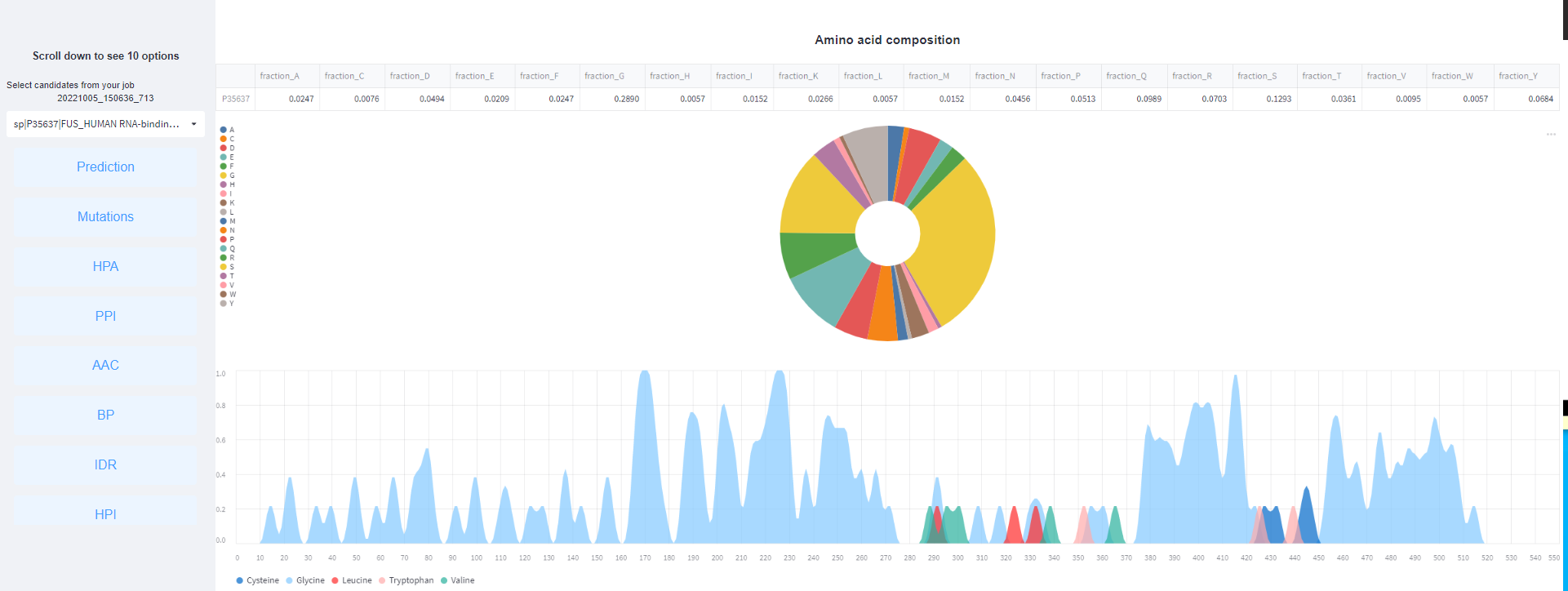
<4>. The results of Amino acid composition.
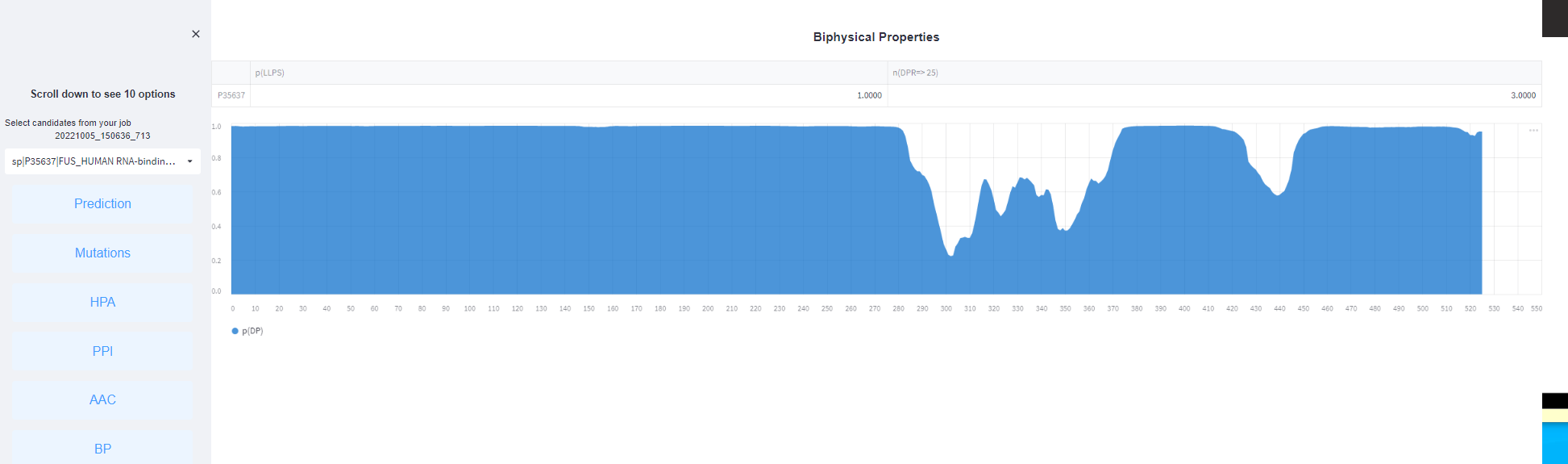
<5>. The results of Biphysical Properties.
<6>. The results of Intrinstically disordered regions.
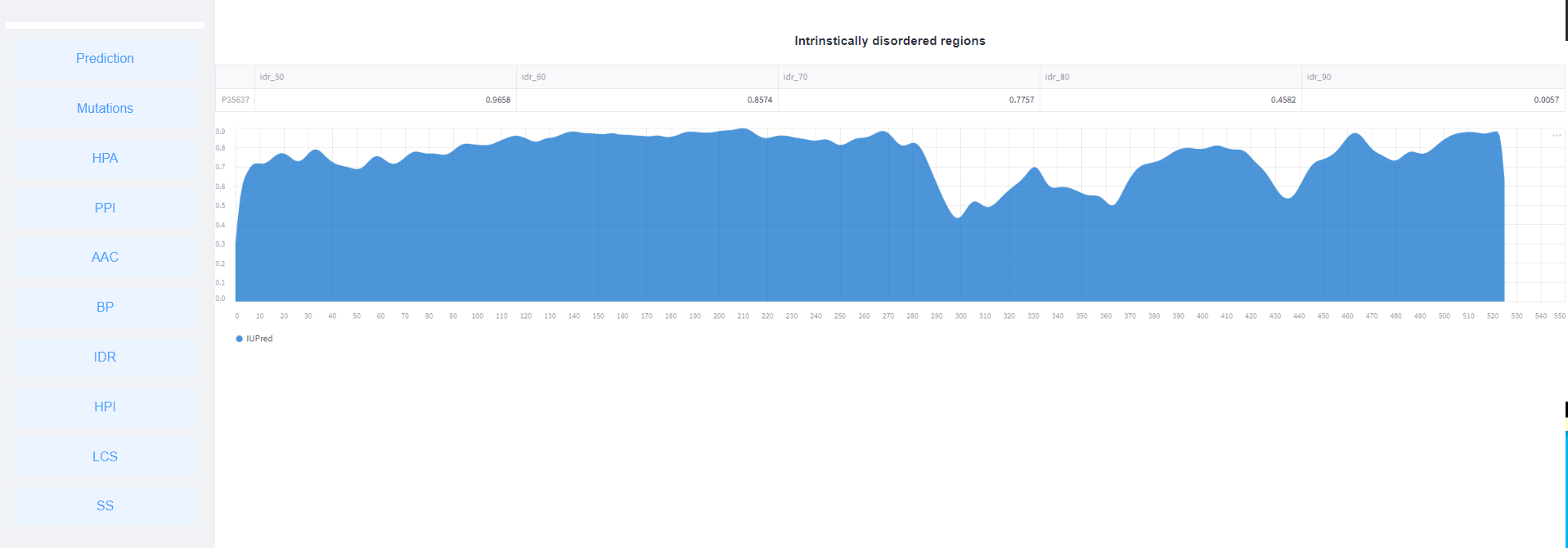
<7>. The results of Hydrophobic index.
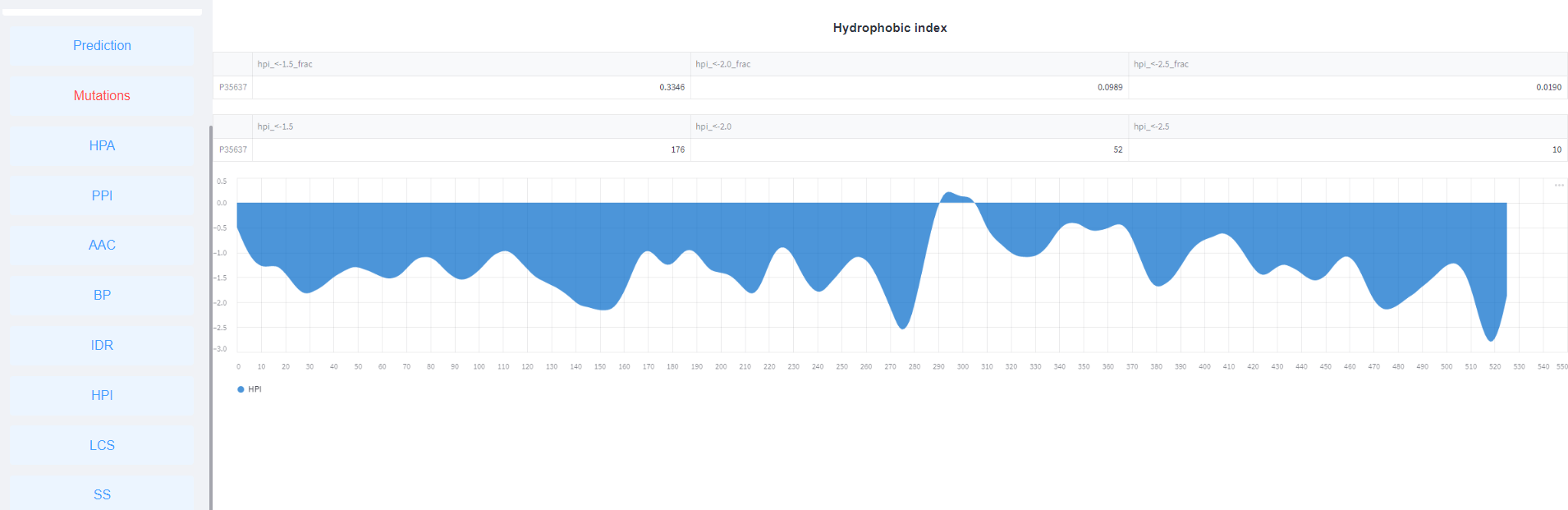
3 . Q: I have a few questions which are not listed above, how can I contact the authors of PULPS?
A: Please contact the major author: Dr. Yaping Guo for details.